【基因检测】基于靶向宏基因组学的耐药性分析
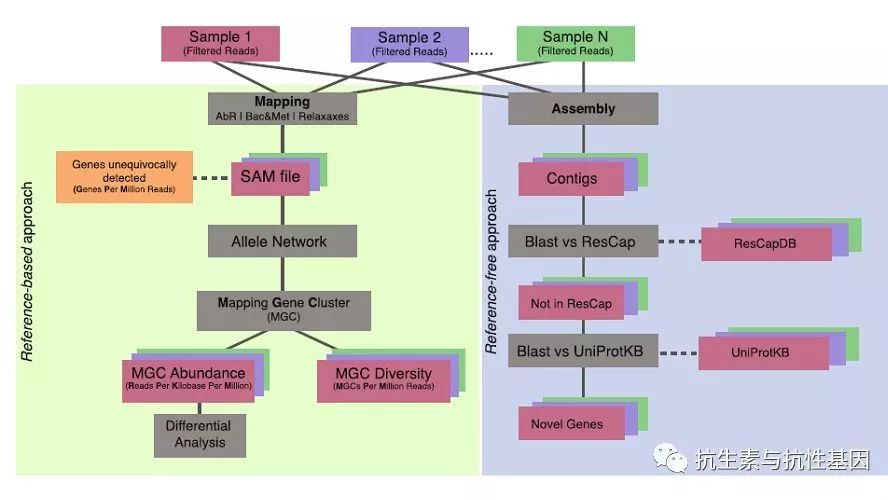
Abstract: Antimicrobial resistance is a major global health challenge. Metagenomics allows analyzing the presence and dynamics of “resistomes” (the ensemble of genes encoding antimicrobial resistance in a given microbiome) in disparate microbial ecosystems. However, the low sensitivity and specificity of available metagenomic methods preclude the detection of minority populations (often present below their detection threshold) and/or the identification of allelic variants that differ in the resulting phenotype. Here, we describe a novel strategy that combines targeted metagenomics using last generation in-solution capture platforms, with novel bioinformatics tools to establish a standardized framework that allows both quantitative and qualitative analyses of resistomes. We developed ResCap, a targeted sequence capture platform based on SeqCapEZ (NimbleGene) technology, which includes probes for 8667 canonical resistance genes (7963 antibiotic resistance genes and 704 genes conferring resistance to metals or biocides), and 2517 relaxase genes (plasmid markers) and 78,600 genes homologous to the previous identified targets (47,806 for antibiotics and 30,794 for biocides or metals). Its performance was compared with metagenomic shotgun sequencing (MSS) for 17 fecal samples (9 humans, 8 swine). ResCap significantly improves MSS to detect “gene abundance” (from 2.0 to 83.2%) and “gene diversity” (26 versus 14.9 genes unequivocally detected per sample per million of reads; the number of reads unequivocally mapped increasing up to 300-fold by using ResCap), which were calculated using novel bioinformatic tools. ResCap also facilitated the analysis of novel genes potentially involved in the resistance to antibiotics, metals, biocides, or any combination thereof. ResCap, the first targeted sequence capture, specifically developed to analyze resistomes, greatly enhances the sensitivity and specificity of available metagenomic methods and offers the possibility to analyze genes related to the selection and transfer of antimicrobial resistance (biocides, heavy metals, plasmids). The model opens the possibility to study other complex microbial systems in which minority populations play a relevant role.
中文摘要:抗微生物药物耐药性是一个重大的全球健康挑战。宏基因组学可以分析不同微生物生态系统中“抗体组合”(在给定的微生物群体中编码抗菌素耐药性的基因组合)的存在和动态。然而,可用的宏基因组学方法的低灵敏度和特异性排除了检测少数群体(通常低于其检测阈值)和/或鉴定所得表型不同的等位基因变体。在这里,我们描述了一种新的战略,结合使用上一代解决方案捕获平台的目标宏基因组学,与新型生物信息学工具,建立一个标准化的框架,允许定性和定量分析的抗体。我们开发了基于SeqCapEZ(NimbleGene)技术的基于SeqCapEZ(NimbleGene)技术的靶向序列捕获平台ResCap,其中包括针对8667个常规抗性基因(7963个抗生素抗性基因和704个赋予金属或杀生物剂抗性的基因)和2517个松弛酶基因(质粒标记)与先前鉴定的靶标同源的78,600个基因(47,806个抗生素和30,794个杀生物剂或金属)。其性能与17个粪便样品(9人,8头猪)的宏基因组鸟枪法测序(MSS)相比较。 ResCap显着改善MSS以检测“基因丰度”(从2.0到83.2%)和“基因多样性”(每百万读数每个样本明确检测到26对比14.9个基因;通过使用明确映射的读数增加达300倍ResCap),这是使用新型生物信息学工具计算的。 ResCap还有助于分析可能参与抗生素,金属,杀生物剂或其任何组合的抗性的新基因。 ResCap是专门开发用于分析抗体组的第一个靶向序列捕获,大大增强了宏基因组方法的灵敏度和特异性,并提供了分析与抗微生物剂抗性(杀生物剂,重金属,质粒)选择和转移有关的基因的可能性。该模型为研究其他复杂的微生物系统开辟了可能性,少数群体在这些微生物系统中起着相关的作用。(Google翻译,未整理,供参考)
DOI: 10.1186/s40168-017-0387-y