并非所有癌症基因突变与乳腺癌风险增加有相关性
前情提要
编者按:虽然随着基因检测水平提高,癌症基因筛查检测项目越来越多,但是并非所有癌症基因突变与乳腺癌风险增加有相关性。那么,在基因检测中的一系列突变中,除了已经非常明确的乳腺癌易感基因和乳腺癌综合征基因之外,究竟哪些基因突变与乳腺癌风险增加有相关性?
2017年4月13日,《美国医学会杂志肿瘤学分册》在线发表梅奥医院、犹他大学、安布瑞基因(免费向全球医学研究组织共享匿名基因数据库的基因测序和遗传咨询公司)、欧文加利福尼亚大学的研究报告,对现有的16个乳腺癌多基因筛查检测项目进行了分析,发现其中一些基因突变确实与乳腺癌有相关性,而另一些过去认为与乳腺癌有相关性的基因突变其实可能与乳腺癌无关。
该病例对照研究对2012年3月15日~2016年6月30日安布瑞基因检测的6万5057例乳腺癌患者(诊断时平均年龄48.5±11.1岁)进行分析。
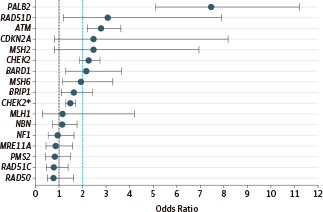
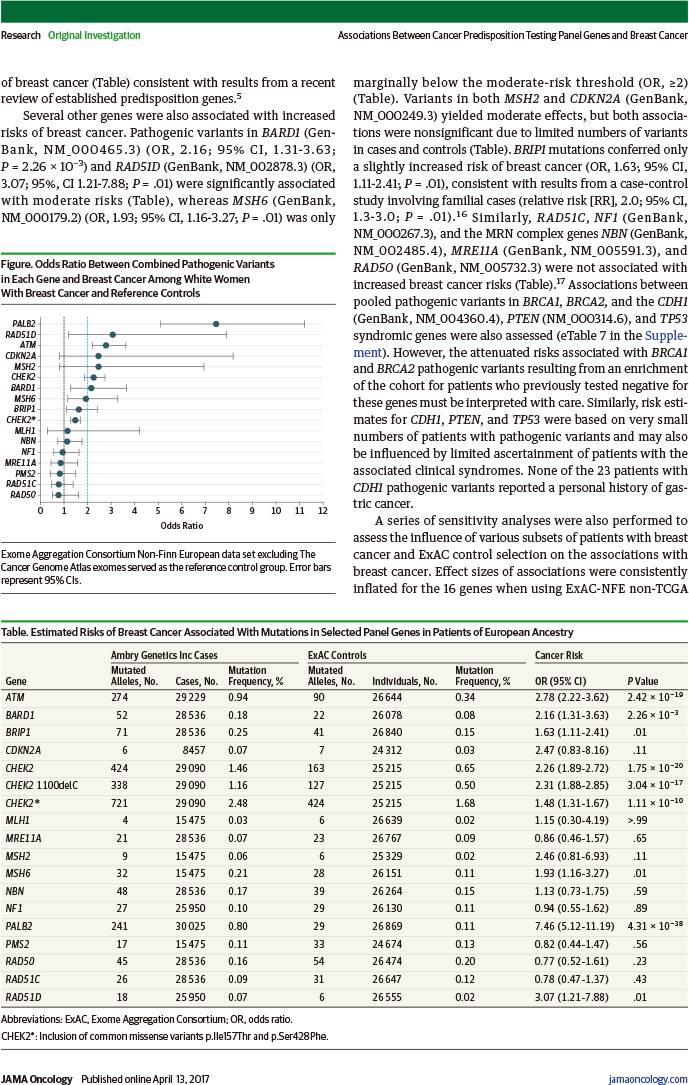
结果发现,除了已经非常明确的乳腺癌易感基因(BRCA1和BRCA2)和乳腺癌综合征基因(CDH1、PTEN、TP53)之外:
-
PALB2基因致病突变与乳腺癌高度风险有相关性(比值比:7.46,95%置信区间:5.12~11.19)。
-
ATM、BARD1、CHEK2、RAD51D突变与乳腺癌中度风险有相关性(比值比:2.78、2.16、1.48、3.07,95%置信区间:2.22~3.62、1.31~3.63、1.31~1.67、1.21~7.88)。
-
BRIP1、RAD51C、MRE11A、RAD50、NBN、MLH1、PMS2、NF1突变与乳腺癌风险增加并无相关性。
因此,虽然多基因检测中的若干致病突变可增加乳腺癌风险,并可使符合癌症监测指征的患者增加,但是其他若干癌症基因突变可能并不会增加乳腺癌风险。
对此,美国福克斯蔡斯癌症中心发表同期述评:乳腺癌风险多基因检测是否应该缩小范围?
该述评认为,将中等风险基因列入多基因检测,可能产生无临床相关性的信息,有时可能对患者及其家属造成误导,故需尽快开展精心设计的研究,以进一步明确低中度风险基因的风险估计,并扩充指南以优化这些患者终生风险的管理。随着将来全外显子组和全基因组检测应用于临床,似乎可以肯定的是,会出现更多的基因突变不确定性。
JAMA Oncol. 2017 Apr 13. [Epub ahead of print]
Associations Between Cancer Predisposition Testing Panel Genes and Breast Cancer.
Fergus J. Couch; Hermela Shimelis; Chunling Hu; Steven N. Hart; Eric C. Polley; Jie Na; Emily Hallberg; Raymond Moore; Abigail Thomas; Jenna Lilyquist; Bingjian Feng; Rachel McFarland; Tina Pesaran; Robert Huether; Holly LaDuca; Elizabeth C. Chao; David E. Goldgar; Jill S. Dolinsky.
Mayo Clinic, Rochester, Minnesota; University of Utah, Salt Lake City; Ambry Genetics Inc, Aliso Viejo, California; University of California-Irvine.
This case-control study evaluates pathogenic variants in panel genes tested in women with breast cancer.
Question: Which genes on hereditary cancer multigene testing panels are associated with high or moderate risks of breast cancer among patients qualifying for clinical genetic testing?
Findings: In a case-control study of 65057 patients with breast cancer, inherited pathogenic variants in PALB2 were associated with high risks of breast cancer, and variants in CHEK2, ATM, BARD1, and RAD51D were associated with moderate risks of breast cancer. Variants in MRE11A, RAD50, NBN, BRIP1, RAD51C, MLH1, and NF1 were not associated with increased risks of breast cancer.
Meaning: Although pathogenic variants in several panel genes confer increased risks of breast cancer and may qualify patients for increased cancer surveillance, variants in several other cancer panel genes may not predispose to breast cancer.
Importance: Germline pathogenic variants in BRCA1 and BRCA2 predispose to an increased lifetime risk of breast cancer. However, the relevance of germline variants in other genes from multigene hereditary cancer testing panels is not well defined.
Objective: To determine the risks of breast cancer associated with germline variants in cancer predisposition genes.
Design, Setting, and Participants: A study population of 65057 patients with breast cancer receiving germline genetic testing of cancer predisposition genes with hereditary cancer multigene panels. Associations between pathogenic variants in non-BRCA1 and non-BRCA2 predisposition genes and breast cancer risk were estimated in a case-control analysis of patients with breast cancer and Exome Aggregation Consortium reference controls. The women underwent testing between March 15, 2012, and June 30, 2016.
Main Outcomes and Measures: Breast cancer risk conferred by pathogenic variants in non-BRCA1 and non-BRCA2 predisposition genes.
Results: The mean (SD) age at diagnosis for the 65 057 women included in the analysis was 48.5 (11.1) years. The frequency of pathogenic variants in 21 panel genes identified in 41611 consecutively tested white women with breast cancer was estimated at 10.2%. After exclusion of BRCA1, BRCA2, and syndromic breast cancer genes (CDH1, PTEN, and TP53), observed pathogenic variants in 5 of 16 genes were associated with high or moderately increased risks of breast cancer: ATM (OR, 2.78; 95% CI, 2.22-3.62), BARD1 (OR, 2.16; 95% CI, 1.31-3.63), CHEK2 (OR, 1.48; 95% CI, 1.31-1.67), PALB2 (OR, 7.46; 95% CI, 5.12-11.19), and RAD51D (OR, 3.07; 95% CI, 1.21-7.88). Conversely, variants in the BRIP1 and RAD51C ovarian cancer risk genes; the MRE11A, RAD50, and NBN MRN complex genes; the MLH1 and PMS2 mismatch repair genes; and NF1 were not associated with increased risks of breast cancer.
Conclusions and Relevance: This study establishes several panel genes as high- and moderate-risk breast cancer genes and provides estimates of breast cancer risk associated with pathogenic variants in these genes among individuals qualifying for clinical genetic testing.
DOI: 10.1001/jamaoncol.2017.0424
JAMA Oncol. 2017 Apr 13. [Epub ahead of print]
Multigene Panel Testing and Breast Cancer Risk: Is It Time to Scale Down?
Elias I. Obeid; Michael J. Hall; Mary B. Daly.
Department of Clinical Genetics, Fox Chase Cancer Center, Philadelphia, Pennsylvania.
The past few years have seen an increase in the adoption of multigene panel tests in the outpatient clinic, providing (in 1 test) comprehensive genetic risk information that, in some cases, can be helpful to guide patients and health care professionals in planning cancer surgery, chemotherapy treatment, screening, and prevention. However, the incorporation of moderate-risk genes into multigene panel tests risks generating information that may not be clinically relevant and at times could be misleading to patients and their families. We are in immediate need of well-designed studies to provide further clarification of risk estimates for low-penetrance and moderate-risk genes, as well as expanded guidelines on how to best manage these risks over the lifetime of the patient. With the anticipated arrival of whole exome and whole genome tests in the clinic in the coming years, what seems most certain is that there is more uncertainty to come.
DOI: 10.1001/jamaoncol.2017.0342